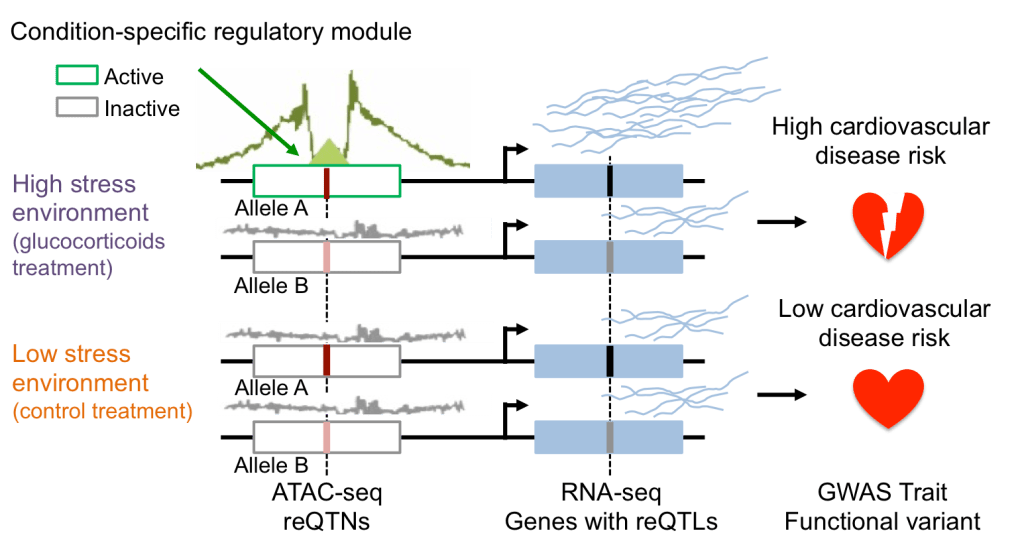
Environmental risk factors are variable in the population and contribute to health disparities. Epidemiological studies have identified several GxE risk factors, for cardiovascular and other diseases, but these findings generally lack functional validation. We characterized and functionally annotated risk variants for cardiovascular diseases in vascular endothelial cells treated with caffeine and other agents, using RNA-seq and ATAC-seq. We detected latent environmental effects and assigned a putative mechanism of action for variants associated with gene expression and coronary artery disease, through disruption of transcription factor binding. Importantly, we found that each treatment may amplify or buffer genetic risk, depending on the particular SNP or gene considered (Findley et al, Genetics, 2019). Using IPSCs and IPSC-derived cardiomyocytes, we demonstrated that context-specific genetic effects on gene regulation are pervasive across cell types and treatments, cannot be captured by large eQTL studies in one single arbitrary context (e.g. GTEx), and explain a significant fraction of heritability and risk for cardiovascular disease and other complex traits (Findley et al, eLife, 2021). We are currently participating in a NIEHS/NHGRI funded initiative to develop in-vitro functional genomics advances for GxE. We are applying the conceptual framework developed to study cellular environment, to explore inter-individual differences in gene-environment interaction responses to endocrine disruptors commonly found in plastics. We are using a combination of RNA-seq, ATAC-seq and MPRA in vascular endothelial and smooth muscle cells, to dissect the regulatory mechanisms underlying these GxE and their contribution to cardiovascular health, through colocalization and transcriptome-wide association study (TWAS) approaches.